








Fluorophores on DNA can be programmably positioned to transfer energy to different degrees through distinct pathways. Our data show distinct energy transfer signatures for each construct.
To control energy transfer for artificial light harvesting technologies or to control the spectroscopic output from a FRET network, it is necessary to engineer a nanoscale structure that controls the relative geometry of the pigments that absorb and transfer energy. DNA's sequence-programmable structure is an ideal platform to design such structures; the relative positions of fluorophores attached to the DNA can be controlled by changing the DNA sequence to produce different spacings or geometries. In particular, custom DNA oligo sequences incorporating doubly-tethered fluorophores such as Cy3 and Cy5 can help align fluorophores with the DNA backbone to limit rotation, providing additional control over the geometric factors that influence energy transfer.
We find that in addition to the DNA-programmed geometry, factors including local sequence and chemical environment strongly influence fluorophore photophysics, providing a large design space with many control knobs to influence the flow of energy through our DNA-fluorophore constructs. We are working to understand this design space and to create structures with distinct energy flow and outputs.

The phycobilisome is the water-soluble primary light harvesting antenna of cyanobacteria. From individual pigment photophysics to protein-protein interactions to structural remodeling, energy flow through the phycobilisome changes dynamically.
Plants, algae, and cyanobacteria have all evolved sophisticated ways to protect themselves from excess sunlight and tune photosynthetic output by removing or rerouting energy. To learn design principles that could mimic these capabilities, we must first understand the underlying dynamic molecular mechanisms that govern natural photoprotection and photoadaptation. A growing body of evidence suggests that dynamic and heterogeneous photophysics in the light-harvesting antennae of cyanobacteria may serve essential biological functions by providing adaptive control over light-harvesting efficiency and energy transfer pathways, allowing cyanobacteria to respond to changing light and environmental conditions with a previously unknown level of sophistication. To study these heterogeneous photophysical states and asynchronous dynamics, which would be obscured by ensemble measurements, it is essential to employ a bottom-up, single-particle approach, and to collect information-rich spectroscopic data that can distinguish photophysical states of phycobilisomes and record the dynamic transitions among them.
We are working to (1) discover the molecular origin of these mechanisms through single-particle spectroscopic measurements and imaging, (2) test the hypothesis that (a) they are induced by changes in incident light or other environmental parameters and that (b) they serve as photoadaptive or photoprotective “switches” and “fuses”, and (3) learn to exert control over these mechanisms. Building this mechanistic bottom-up view of the intrinsic photodynamic sophistication of phycobilisomes is essential for extending our biological understanding of efficient and adaptive solar energy capture and transport in photosynthesis and will also provide useful guidance and inspiration for rational design of photoadaptive synthetic light harvesting systems and other green technologies.

Upon heat shock, protein condensates form inside cells, which must be later de-aggregated by chaperone proteins in order to resume normal function.
The cell environment drives cell biology; cells must constantly sense and respond to changing chemical and physical signals in their local environment in order to survive. One such response occurs when the environmental temperature delivers a “heat shock” to the cell. Upon heat shock, many proteins aggregate to form condensates, and transcription of chaperone proteins is initiated. These chaperones are responsible for disaggregation of protein condensates after heat shock, eventually restoring normal function to the cell. Although the heat shock response is well-conserved across species, molecular-scale understanding of how condensates and heat shock proteins interact, and of the dynamic interactions among them, are not known.
We are using single-molecule fluorescence imaging to directly observe the action of heat shock proteins as they regulate heat-induced protein aggregates. By monitoring changes in the number and locations of these proteins, and detecting their proximity with FRET, we can learn the fundamental stoichiometry, location, and dynamics of the molecular mechanisms enabling this critical stress response.

Rich spectroscopic information is present in fluorescence signals, particularly when energy transfer occurs.
Spectroscopic fluorescence data reveal intrinsic and information-rich features of fluorescence that are overlooked by traditional microscopy. Fluorophores are (usually) simple two-level electronic systems that require different characteristic amounts of time to absorb and emit photons, corresponding to fluorescence lifetime. Fluorescent photons are emitted by transitions at characteristic energies, corresponding to emission wavelength. Fluorophores are antenna-like dipoles, so emitted photons have particular orientations, or anisotropy (polarization). And because some fluorophores are more efficient than others, they can have greater brightness.
This information provides insight into the nanoscale environment of a fluorophore, as well as its position relative to other molecules, during dynamic events occurring between photon absorption and emission. With these additional insights, spectroscopic fluorescence approaches vastly enhance standard biological imaging and detection applications, particularly at the single-molecule level.
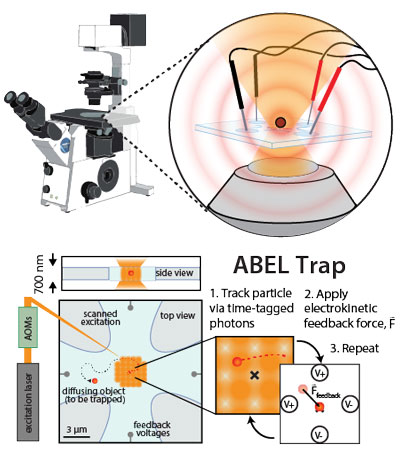
The ABEL trap holds a single molecule (as small as a single fluorophore) in free solution using closed-loop electrical feedback.
Brownian motion of small particles in free solution can impede or even prohibit optical studies of nanometer-sized objects, such as single biomolecules, because such objects quickly diffuse away from the observation region. Anti-Brownian traps address this challenge by partially suppressing Brownian motion: the position of a single particle is monitored, and active feedback is used to apply forces that directly counteract the observed displacements. This process confines the particle to a small region of interest (ROI), enabling extended study without surface attachment or encapsulation, both of which could perturb the particle’s behavior.
We frequently use our custom-built Anti-Brownian ELectrokinetic (ABEL) trap (based on the original design by Cohen and Moerner, 2006) to hold molecules or particles in place while we take high-precision measurements. Thanks to the long observation times and isotropic view of the sample enabled by our trap, dynamics and/or transitions among many distinct emissive states can be directly visualized. Because movements of the particle are recorded with and without electrical feedback, trap data can also be used to determine the diffusion coefficients and electrophoretic mobility of samples in real time. In addition to our ongoing projects, we are presently working to expand the capabilities of the trap to work with different types of samples, and in different contexts, to enable better actuation at the nanoscale.